![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
Archives
& Museum Informatics
158 Lee Avenue
Toronto, Ontario
M4E 2P3 Canada
info@ archimuse.com
www.archimuse.com
|
|
Search A&MI |
Join
our Mailing List.
Privacy.
Published: March 15, 2001.

Digital Embryo Library and Collaboratory Tools
Mike Doyle, Eolas Technologies, Elizabeth Lockett, National Museum of Health & Medicine, Arcot Rajasekar, University of California, San Diego, USA
Abstract
Officially titled "Human Embryology Digital Library and Collaboratory Support Tools," this project is part of the Next Generation Internet Initiative and is funded by the National Library of Medicine, which is part of the National Institutes of Health. The project's purpose is to demonstrate how leading-edge information technologies in computation, visualization, collaboration, and networking can expand capabilities in science and medicine for developmental studies, clinical work and teaching. It also shows how old collections are being made useful with modern technology. Data for the project comes from the Carnegie Human Embryology Collection at the National Museum of Health and Medicine of the Armed Forces Institute of Pathology in Washington, DC. The project will draw from carefully prepared serially-sectioned specimens acquired between the 1890s and the 1930s.While the project primary focus is on providing a capability for medical professionals and biological scientists to communicate detailed information about development of the human embryo in a visual form, it also contains a component for K-12 and the general public to access and view the HDAC collections. For researchers, the project technical team will develop a network of medical collaboration workstations, using high-performance off-the-shelf networked computer systems combined with advanced software for collaboration and medical visualization. The workstations will be installed at eight project locations and interconnected over high performance networks. As a result, physicians and others will be able to visualize and manipulate high-resolution image data collaboratively for diagnoses, clinical case management, and medical education. For the general public an interface using standard Internet technologies will be provided to view lower resolution sets of image data, access educational information, lab tools and animations.The project plans to demonstrate the network of collaborative visualization workstations in three advanced applications:
- The Annotation and Modeling application will create an archive of tagged image data for visualization, where every picture element can be associated with any number of different labels, for example the organ system to which it belongs, the structure and tissue types represented, the specimen to which it belongs, etc. This will provide the basic archive that the other two applications will use
- The Embryology Education application will make the visualization tools available for medical student use, and also will create animations of embryo organ system development. Heart, lungs and perineal region will be modeled
- The Clinical Management Planning application will make the data available in visual form so that a geographically distributed group of physicians can look at it simultaneously, manipulate it, and discuss it.
History
Who is AFIP, NMHM and HDAC
The National Museum of Health and Medicine, formerly the Army Medical Museum, The National Museum of Health and Medicine was established during the Civil War as the Army Medical Museum, a center for the collection of specimens for research in military medicine and surgery. In 1862, Surgeon General William Hammond directed medical officers in the field to collect "specimens of morbid anatomy . . . together with projectiles and foreign bodies removed" and to forward them to the newly founded museum for study.
By World War II, research at the Museum focused increasingly on pathology; in 1946, the Museum became a division of the new Army Institute of Pathology (AIP), which became the Armed Forces Institute of Pathology (AFIP), in 1949. The Museum's library and part of its archives were transferred to the National Library of Medicine when it was created in 1956. The Army Medical Museum became the National Museum of Health and Medicine in 1989.
As a division of the Museum, the Human Developmental Anatomy Center was the brainchild of the then Assistant Director of the museum, Adrianne Noe. The Carnegie Institute of Washington, from1890 to 1955 amassed a collection of embryos taken upon mother's autopsy, miscarriage, or therapeutic termination of pregnancy. When the Institute's Department of Embryology began its move into Cellular Biology and wanted to divest itself of its physical specimens, the Institute was persuaded that the best home for their collection was the National Museum of Health and Medicine. There the collection could become a widely available resource for research and education. With the addition of several other important American embryology collections, the Human Developmental Anatomy Center now houses material from 10,000+ individuals, the largest collection of normal human embryology in the US.
Current Project
Officially titled "Human Embryology Digital Library and Collaboratory Support Tools,"(or Visible Embryo for short) this project is part of the Next Generation Internet Initiative and is funded by the National Library of Medicine, which is part of the National Institutes of Health. The project's purpose is to demonstrate how leading-edge information technologies in computation, visualization, collaboration, and networking can expand capabilities in science and medicine for developmental studies, clinical work and teaching. The Visible Embryo project will draw from carefully prepared serially sectioned specimens acquired between the 1890s and the 1930s. While the project focuses on providing a capability for medical professionals and biological scientists to communicate detailed information about development of the human embryo in a visual form, it also contains a component for K-12 and the general public to access and view the collections. For researchers, the project technical team will develop a network of medical collaboration workstations, using high-performance off-the-shelf networked computer systems combined with advanced software for collaboration and medical visualization. The workstations will be installed at eight project locations and interconnected over high performance networks. Consequently, physicians and others will be able to visualize and manipulate high-resolution image data collaboratively for diagnoses, clinical case management, and medical education. For the general public an interface using standard Internet technologies will be provided to view lower resolution sets of image data, access educational information, lab tools and animations.
Technical participants in the "Visible Embryo" project are:
- George Mason University of Fairfax, Virginia, which
has overall responsibility for the project, and will provide collaboration
technology.
- Armed Forces Institute of Pathology of Washington, DC, is the collaborating medical group that holds the Carnegie Collection at the National Museum of Health and Medicine. It will be responsible for automating and capturing digital image data, aiding in the development of the knowledge management system (database), and visualization system.
- Eolas Technologies, Inc. of Chicago, Illinois, will be responsible for software development including the advanced visualization system, information mapping system, knowledge management system, and groupware environment.
- Lawrence Livermore National Laboratory of Livermore, California will be responsible for arranging access to high performance networks, integrating the applications with the network, and doing network performance monitoring and tuning.
- San Diego Supercomputer Center operated by the University of California at San Diego, will be responsible for maintaining the medical image database, which will be one of the largest ever created, and for providing supporting software.
- The collaborating medical groups in the project will participate in each of the applications. Each group will have a leading role in one application:
- Johns Hopkins University Medical School of Baltimore, Maryland, will have a leading role in the Clinical Management Planning application, and will be responsible for aspects of embryology dealing with the perineal region in all three applications.
- Oregon Health Science University of Portland, Oregon, will have a leading role in the Annotation and Modeling application, and will be responsible for aspects of embryology dealing with the heart and lungs in all three applications.
- University of Illinois at Chicago Illinois will have a leading role in the Embryology Education application, and will be responsible for aspects of embryology dealing with the upper digestive tract in all three applications.
- Armed Forces Institute of Pathology of Washington, DC, is the collaborating medical group that holds the Carnegie Collection at the National Museum of Health and Medicine. It will be responsible for automating and capturing digital image data, aiding in the development of the knowledge management system (database), and visualization system.
Developing database
During the first proof of concept, users of the collection from various fields were solicited for information about how they used the data, how they accessed the data and what they would like to see in applications and visualizations of the data. This information provided the basis for the design of the database.
To try to accommodate all the various applications and uses of the data was something of a logistical challenge. The database would be required to accommodate collection management applications, storage of images and image information, and storage of information generated by researchers while using collaborative tools.
Data Definition Tables
After reviewing several data structures from scientific organizations and museums, and identifying the field types required for most of the various applications the database would need to support, Data Definition Tables were generated. DTD's for collections management and automated microscopy were generated by HDAC. DTD's for the collaborative tools were written by SDSC in conjunction with Eolas Technologies.
With the guidance of the San Diego Super Computing Center, HDAC staff started by modifying some basic collections management type Data Definition Tables created for AMICO by SDSC. By looking at those examples and the tables already in existence at HDAC, a set of ddt's were developed to accommodate collections management activities, ongoing imaging activities of various types and needs of the users. One of the first considerations in developing the metadata catalogue was whether to use Dublin Core metadata. Several of the basic Dublin Core fields such as Object Title/Name and Materials and Techniques Description, were used to try to provide some useful connectivity should it be requested in the future. Other basic tables for verification had to be identified and created. Metadata as it currently existed had to be converted and put into new table structures. This usually meant moving a field from its current table to the new table. Occasionally, data within a field had to be split apart into to new fields. This was usually done to try to simplify information in fields to one type information. Fields, which were duplicated in several tables, were consolidated by creating one table to contain that data and link keys in other tables to it.
Verification Language and Decisions about language
After some searching and discussion, the international standard Nomina Anatomica (formerly Terminologica Anatomica) was chosen for adult anatomical nomenclature, Nomina Embryologica, still in development, was chosen for developing anatomy. Both these thesauruses are juried by international panels and have terms in Latin and English. SNOWMED was chosen for pathology, as it is the most common standard used by hospitals and clinicians, and Art and Architecture for collection management and artifact terms. Investigations of the UMLS system developed by the National Library of Medicine are still ongoing.
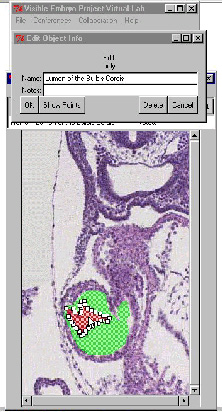
Oregon Health Sciences University researchers will be outlining and labeling structures on embryo section images. Working with OHSU will be Dr. Cornilius Rosse who developed the hierarchical naming system for the Digital Anatomist Program at the National Library of Medicine, and Dr. Raymond Gasser a prominent embryologist on the Nomina Embryologica panel. These annotations will provide researchers and viewers a set of section images from embryos with all the prominent structures visible outlined and labeled.
Developing the automated imaging process
Hardware
SONY DTK5 digital video cameras attached to Nikon E800 microscopes with plan apo lenses. 2000X2000 resolution images are 15mb each. Ludl stages allow for automatic 3 dimensional stage movement. Dell computers with high-end video cards and extra memory provide local storage, and microscope control. A Silicon Graphics Origin 200 with a 200 GB RAID works as a local data server and short-term storage.
Fig2: Workflow and software development
Off the shelf products form the core of tools used to image and visualize data. ImagePro (Media Cybernetics) provides basic RS-232 control in support of stage manipulation. This includes client development API to script end-user modifications written by graduate student from George Mason University, Dave Brooks. Imagewhere software from Where?media ltd. Was modified to create several watchdogs to help automate the archiving process and provide a tool to view data via regular Internet.
Current imaging procedure was reviewed to see where automated processes could be added. After review, a decision was made to dedicate one microscope to 2x capture and 2 scopes to 20x capture. This eliminated problems in variations due to adjusting condensers and filters and lenses. All slide capture begins with calibration of the 2X and 20X scopes by using travel limits of the stage, x0,y0 in the upper left. A spectral calibration is made and stored as metadata, and on 20x capture a verification of overlap between tiles is done.
On the 2X capture scope, software functions are exercised to automatically capture tiled images of the cover slipped area of a 2X3 slide. These images provide the spatial map for the 20x capture which follows. On the 20X scopes, Imagepro calls up the 2X spatial map of the slide. On this image the user must draw boxes called the Regions Of Interest (ROI) around areas containing data. These will be the areas, which are captured at 20X. These coordinates are stored as metadata, incorporated into the XML document later.
Collection and storage of data
Tissue section images form the largest portion of the data collected and served by this project.
Tiles - 15 MB per tile
Sections - up to 100 tiles per section - 1.5 GB
Embryo - up to 700 sections per embryo - 1 TB
Collected data is stored initially as a lossless TIFF image. Metadata such as session information, information about the lens and filters, regions of interest and XY coordinates necessary for reconstruction of tiled images is transferred via software generated XML documents. Imagewhere watchdogs, internal scripts that monitor specified directories for new files, move images from capture stations to a UNIX based RAID large enough to hold 3 days of capture information, ~200 GB. Using conversion tools resident on the UNIX machine, smaller lossey JPEG sets are created of the 2X maps for use in regular net viewing tools. Imagewhere archives a set of metadata, which stays resident on a local server, as well as a set of the smaller lossy images. This provides search and visualization capability for users via html. Images and metadata are grabbed by other archiving watchdogs that automatically push images and metadata to the San Diego Super Computing (SDSC) facility where 10 terabytes of on-line storage are available for the project. Where?media's interface also provides a tool for visual checks of the data before and during transfer to SDSC, as well as the broad internet user interface.
SDSC serves and controls data with the Storage Resource Broker (SRB). This tool acts as an intermediary, locating data through out a distributed system for any requestor by giving data URL and updating that URL association on the fly so if data is moved to another location, the SRB will keep track and automatically update the database and metadata catalogue with the new URL.
Educational tools
The University of Illinois Chicago Medical School, working with the Biomedical Visualization Department, are creating embryo models using the images collected from the project. These are being combined with other technologies to create on-line courses in embryology, which are being incorporated into the University curriculum. These tools will be evaluated and modified over the 3-year project.
Collaborative Tools
The first of the collaborative tools to be developed was the incorporation and modification of on-line conferencing tools by George Mason University. Using M-Bone technology video and whiteboard tools provide ways for remote collaborators to meet bi-weekly to discuss the project. The second tool underdevelopment by Eolas Technologies has been an on-line annotation system that will allow researchers in OHSU to outline and label structures on the embryo section images. These annotations and labels are saved into the database and become part of the information that all researchers and students are able to query and access on-line. The outline of the structures and organ systems become the basis for 3-D computer models which are created "on-fly" by SDSC servers as requested. The labels associated with a structure in 2-D stay associated in the 3-D map by using associations in the hierarchical naming system.
Other interactive tools for the clinical teams are in very early stages of development and will be produced in later stages of the project.
Future
The neuro anatomical collections in the museum has many similar requirements to those of HDAC. The current initiative will translate well to this other department and provide many useful tools that are not currently available to its researchers. Use of the labeled models for gene mapping and gene expression is currently under investigation.